-Search query
-Search result
Showing 1 - 50 of 55 items for (author: zhu & dj)

EMDB-34227: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 1
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

EMDB-34229: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 4
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

EMDB-34230: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 2
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

EMDB-34235: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 3
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

EMDB-34236: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 5
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF
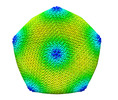
EMDB-34251: 
Singapore Grouper Iridovirus
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

EMDB-34815: 
One asymmetric unit of Singapore grouper iridovirus capsid
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

EMDB-26636: 
E.coli RNaseP Holoenzyme with Mg2+
Method: single particle / : Huang W, Taylor DJ

EMDB-26637: 
E.coli RNaseP Holoenzyme with Mg2+
Method: single particle / : Huang W, Taylor DJ

EMDB-26638: 
E.coli RNaseP Holoenzyme with Mg2+
Method: single particle / : Huang W, Taylor DJ

EMDB-26640: 
E.coli RNaseP Holoenzyme with Mg2+
Method: single particle / : Huang W, Taylor DJ

EMDB-26522: 
SARS-CoV-2 6P Mut7 in complex with K398.25 Fab
Method: single particle / : Lee WH, Torres JL, Ward AB

EMDB-26523: 
SARS-CoV-1 in complex with K398.25 Fab
Method: single particle / : Lee WH, Torres JL, Ward AB

EMDB-26524: 
SARS-CoV-2 6P Mut7 in complex with K398.16 Fab
Method: single particle / : Lee WH, Torres JL, Ward AB
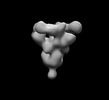
EMDB-26525: 
SARS-CoV-2 6P Mut7 in complex with K398.16 Fab (3 bound)
Method: single particle / : Lee WH, Torres JL, Ward AB

EMDB-26526: 
SARS-CoV-1 in complex with K398.16 Fab
Method: single particle / : Lee WH, Torres JL, Ward AB

EMDB-26527: 
SARS-CoV-2 6P Mut7 in complex with K288.2 Fab
Method: single particle / : Lee WH, Torres JL, Ward AB

EMDB-26528: 
SARS-CoV-2 6P Mut7 in complex with K398.8 Fab
Method: single particle / : Lee WH, Torres JL, Ward AB

EMDB-26529: 
SARS-CoV-2 6P Mut7 in complex with K398.8 Fab (2 bound)
Method: single particle / : Lee WH, Torres JL, Ward AB

EMDB-26530: 
SARS-CoV-2 6P Mut7 in complex with K398.18 Fab
Method: single particle / : Lee WH, Torres JL, Ward AB

EMDB-26531: 
SARS-CoV-2 6P Mut7 in complex with K398.18 Fabs (2 bound)
Method: single particle / : Lee WH, Torres JL, Ward AB

EMDB-26532: 
SARS-CoV-2 6P Mut7 in complex with K398.22 Fab
Method: single particle / : Lee WH, Torres JL, Ward AB
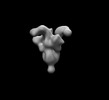
EMDB-26533: 
SARS-CoV-2 6P Mut7 in complex with K398.22 Fab (2 bound)
Method: single particle / : Lee WH, Torres JL, Ward AB

EMDB-26534: 
SARS-CoV-1 in complex with K398.8 Fab
Method: single particle / : Lee WH, Torres JL, Ward AB

EMDB-26535: 
SARS-CoV-1 in complex with K398.8 Fab (2 bound)
Method: single particle / : Lee WH, Torres JL, Ward AB

EMDB-26536: 
SARS-CoV-1 in complex with K398.18 Fab (2 bound)
Method: single particle / : Lee WH, Torres JL, Ward AB

EMDB-26537: 
SARS-CoV-1 in complex with K398.18 Fab (3 bound)
Method: single particle / : Lee WH, Torres JL, Ward AB

EMDB-26538: 
SARS-CoV-1 in complex with K288.2 Fab
Method: single particle / : Lee WH, Torres JL, Ward AB

EMDB-26539: 
SARS-CoV-1 in complex with K398.22 Fab
Method: single particle / : Lee WH, Torres JL, Ward AB

EMDB-32611: 
Structure of Coxsackievirus A10 with Hybrid electron counting at 200 kV
Method: single particle / : Zhu DJ, Zhang XZ

EMDB-32600: 
Structure of Coxsackievirus A10 for critical dose measurement at 120 kV
Method: single particle / : Zhu DJ, Zhang XZ

EMDB-32601: 
Structure of Coxsackievirus A10 for critical dose measurement at 160 kV
Method: single particle / : Zhu DJ, Zhang XZ

EMDB-32602: 
Structure of Coxsackievirus A10 for critical dose measurement at 300 kV
Method: single particle / : Zhu DJ, Zhang XZ

EMDB-32603: 
Structure of Coxsackievirus A10 with Hybrid electron counting at 120 kV
Method: single particle / : Zhu DJ, Zhang XZ

EMDB-32604: 
Structure of Coxsackievirus A10 with MCF electron counting at 120 kV
Method: single particle / : Zhu DJ, Zhang XZ

EMDB-32605: 
Structure of Coxsackievirus A10 with WPF electron counting at 120 kV
Method: single particle / : Zhu DJ, Zhang XZ

EMDB-32606: 
Structure of Coxsackievirus A10 with MCF electron counting and large-sized clusters at 120 kV
Method: single particle / : Zhu DJ, Zhang XZ

EMDB-32607: 
Structure of Coxsackievirus A10 with PVF electron counting and large-sized clusters at 120 kV
Method: single particle / : Zhu DJ, Zhang XZ

EMDB-32608: 
Structure of Coxsackievirus A10 with PCA electron counting and large-sized clusters at 120 kV
Method: single particle / : Zhu DJ, Zhang XZ

EMDB-32609: 
Structure of Coxsackievirus A10 with MCF electron counting at 200 kV
Method: single particle / : Zhu DJ, Zhang XZ

EMDB-32610: 
Structure of apo-ferritin with PCA electron counting at 300 kV
Method: single particle / : Zhu DJ, Zhang XZ

EMDB-32612: 
Structure of apo-ferritin with Hybrid electron counting at 120 kV
Method: single particle / : Zhu DJ, Zhang XZ

EMDB-32613: 
Structure of apo-ferritin with MCF electron counting at 120 kV
Method: single particle / : Zhu DJ, Zhang XZ

EMDB-11978: 
Nanobody E bound to Spike-RBD in a localized reconstruction
Method: single particle / : Hallberg BM, Das H

EMDB-11981: 
SARS-CoV-spike bound to two neutralising nanobodies
Method: single particle / : Hallberg BM, Das H

EMDB-11980: 
SARS-CoV-spike RBD bound to two neutralising nanobodies.
Method: single particle / : Hallberg BM, Das H

EMDB-23018: 
SARS-CoV-2 spike in complex with nanobodies E
Method: single particle / : Hallberg BM, Das H

EMDB-0535: 
Cryo-EM Reconstruction of Protease-Activateable Adeno-Associated Virus 9 (AAV9-L001)
Method: single particle / : Bennett AB, Agbandje-Mckenna M

PDB-6nxe: 
Cryo-EM Reconstruction of Protease-Activateable Adeno-Associated Virus 9 (AAV9-L001)
Method: single particle / : Bennett AB, Agbandje-Mckenna M

EMDB-6976: 
Structure of the Herpes simplex virus type 2 C-capsid with capsid-vertex-specific component
Method: single particle / : Wang JL, Yuan S, Zhu DJ, Tang H, Wang N, Chen WY, Gao Q, Li YH, Wang JZ, Liu HR, Zhang XZ, Rao ZH, Wang XX
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
